FLASHDECONV 2.0 BETA+, FINALLY WITH A GUI!
Finally a GUI is here. You can find the GUI command in [OpenMS path]/bin folder. Go to [OpenMS path]/bin and run FLASHDeconvWizard! FLASHDeconv 2.0 beta+ works for MS1 and MS2 spectral deconvolution and feature deconvolution. It supports various output formats (e.g., *.tsv, *.mzML, *.msalign, and *.feature). FLASHDeconv 2.0 stable version will be officially integrated in OpenMS 2.7.0 released in near future. FLASHDeconv 2.0 beta+ also supports TopPIC identification better than the previous version, by generating all msalign and feature files for TopPIC inputs. We also added spectral merging function to support QTOF dataset analysis and NativeMS dataset analysis.
Changes:
- FLASHDeconvWizard (GUI) is added!
- FLASHIda support (-in_log option)
- We no longer recommend profile mode spectra. Peak picked spectra (by MSConvert vendor provided peak picking) are recommended as inputs.
- merging_method option is introduced to merge or average MS2 spectra.
- use_ensemble_spectrum option has been removed (replaced by -merging_method).
- target_mass option is added to perform targeted deconvolution (deconvolution quality control is relaxed for target masses) – target_sequence or proteoform option will be soon added.
- min_precursor_snr option is introduced that (currently) only affects msalign and feature files for TopPIC.
- out_topFD_feature option is introduced that outputs feature file for TopPIC. In TopPIC, no need to use -x option with this feature file input.
- Quality measure score (QScore) is added per each deconvolved mass in spectral deconvolution results. QScore is the probability that a mass is identified, learned by a logistic regression (related publication will be added here). Note that it is the probability that the mass is “identified” not “correct.”
- Both MS1 and MS2 deconvolution have been extensively improved (tested by proteoform ID sensitivity, coupled with TopPIC).
- Works well for both centroid and profile spectra. In particular for MS2, centroid spectra should be used.
- Support negative charges (set by -Algorithm:min_charge and -Algorithm:max_charge parameters; see below).
- Parameter set is redefined (see below).
- Batch execution is not supported for FLASHDeconv binary. Separate batch files will be prepared soon.
- Deconvolved spectra may be output in mzml format (-out_mzml [mzml file]).
- Deconvolved MS1 spectra may be output in Promex format (-out_promex [ms1ft file]).
- Deconvolved MS1/2 spectra may be output in TopFD format (-out_topFD [msalign file per MS level]).
- Deconvolved MS1/2 features may be output in TopFD format (-out_topFD_feature [feature file per MS level]).
- Effective harmonic artifact elimination in mass dimension effectively reduces false negatives while keeping true positives.
Under development
- Proforma 2.0 support (-target_seq option)
- Deep learning based deconvolution quality measure
- QScore training interface
- Parameter set for different protocols (e.g., Native-MS, HighRes TDP, …)
- Merge into OpenMS 3.0
Installation
FLASHDeconv installation files (OpenMS-2.x.0-HEAD-, for windows *.exe, for mac *.dmg, and for linux .deb) and source code (-src.tar.gz) are found in here. For the latest version, go to the bottom side of the page and select the most recent installation file.
Parameters
FLASHDeconv basic parameters are found by simply running FLASHDeconv. Only -in and -out are mandatory. FLASH advanced parameters are found by running FLASHDeconv –helphelp. FLASHDeconv parameters have six categories: FLASHDeconv tool parameters, FLASHDeconv algorithm parameters, Spectral deconvolution parameters, Feature tracing parameters, Isobaric quantification parameters and Tagger parameters. Firstly the basic parameters in each category are described, and then the advanced ones are explained.
FLASHDeconv tool parameters:
| Parameter | Description |
|---|---|
| in |
Input file (mzML) (valid formats: ‘mzML’) |
| out |
Default output tsv file containing deconvolved features (valid formats: ‘tsv’) |
| out_spec1 |
Output tsv file containing deconvolved MS1 spectra. Likewise, use -out_spec2, …, -out_spec4 to specify tsv files for MS2, …, MS4. (valid formats: ‘tsv’) |
| out_spec2 |
Output tsv file containing deconvolved MS2 spectra. (valid formats: ‘tsv’) |
| out_spec3 |
Output tsv file containing deconvolved MS3 spectra. (valid formats: ‘tsv’) |
| out_spec4 |
Output tsv file containing deconvolved MS4 spectra. (valid formats: ‘tsv’) |
| out_mzml |
Output mzml file containing deconvolved spectra (of all MS levels) (valid formats: ‘mzML’) |
| out_quant |
Output tsv file containing isobaric quantification results for MS2 only (valid formats: ‘tsv’) |
| out_annotated_mzml |
Output mzml file containing annotated spectra. For each annotated peak, monoisotopic mass, charge, and isotope index are stored as meta data. Unannotated peaks are also copied as well without meta data. (valid formats: ‘mzML’) |
| out_msalign1 |
Output msalign (topFD and ProMex compatible) file containing MS1 deconvolved spectra. Likewise, use -out_msalign2 for MS2 spectra. The file names for MS1 and MS2 should end with ms1.msalign and ms2.msalgin respectively to be able to be recognized by TopPIC GUI. (valid formats: ‘msalign’) |
| out_msalign2 |
Output msalign (topFD and ProMex compatible) file containing MS2 deconvolved spectra. The file name should end with ms2.msalign to be able to be recognized by TopPIC GUI. (valid formats: ‘msalign’) |
| out_feature1 |
Output feature (topFD compatible) file containing MS1 deconvolved features. Likewise, use -out_feature2 for MS2 features. The MS1 and MS2 feature files are necessary for TopPIC feature intensity output. (valid formats: ‘feature’) |
| out_feature2 |
Output feature (topFD compatible) file containing MS2 deconvolved features. The MS1 and MS2 feature files are necessary for TopPIC feature intensity output. (valid formats: ‘feature’) |
| keep_empty_out | If set, empty output files (e.g., *.tsv file when no feature was generated) are kept. |
| mzml_mass_charge <0: uncharged 1: +1 charged -1: -1 charged> | Charge state of deconvolved masses in mzml output (specified by out_mzml) (default: ‘0’) (min: ‘-1’ max: ‘1’) |
| write_detail | To write peak information per deconvolved mass in detail or not in tsv files for deconvolved spectra. If set to 1, all peak information (m/z, intensity, charge and isotope index) per mass is reported. |
| precursor_snr |
Precursor SNR threshold for TopFD MS2 msalign tsv files. (default: ‘1.0’) |
| min_mz <m/z value> | If set to positive value, minimum m/z to deconvolve. (default: ‘-1.0’) |
| max_mz <m/z value> | If set to positive value, maximum m/z to deconvolve. (default: ‘-1.0’) |
| min_rt | If set to positive value, minimum RT (in second) to deconvolve. (default: ‘-1.0’) |
| max_rt | If set to positive value, maximum RT (in second) to deconvolve. (default:'-1.0') |
| ini |
Use the given TOPP INI file |
| log |
Name of log file (created only when specified) |
| instance |
Instance number for the TOPP INI file (default: ‘1’) |
| debug |
Sets the debug level (default: ‘0’) |
| threads |
Sets the number of threads allowed to be used by the TOPP tool (default: ‘1’) |
| write_ini |
Writes the default configuration file |
| write_ctd <out_dir> | Writes the common tool description file(s) (Toolname(s).ctd) to <out_dir> |
| write_nested_cwl <out_dir> | Writes the Common Workflow Language file(s) (Toolname(s).cwl) to <out_dir> |
| write_cwl <out_dir> | Writes the Common Workflow Language file(s) (Toolname(s).cwl) to <out_dir>, but enforce a flat parameter hierarchy |
| write_nested_json <out_dir> | Writes the default configuration file |
| write_json <out_dir> | Writes the default configuration file, but compatible to the flat hierarchy |
| no_progress | Disables progress logging to command line |
| force | Overrides tool-specific checks |
| test | Enables the test mode (needed for internal use only) |
| -help | Shows options |
| -helphelp | Shows all options (including advanced) |
FLASHDeconv algorithm parameters (with prefix FD: )
| parameter | Description |
|---|---|
| FD:ida_log |
Log file generated by FLASHIda (IDA*.log). Only needed for coupling with FLASHIda acquisition |
| FD:report_FDR | Report qvalues (roughly, point-wise FDR) for deconvolved masses. Decoy masses to calculate qvalues and FDR are also reported. Beta version. |
| FD:allowed_isotope_error |
Allowed isotope index error for decoy and FDR report. If it is set to 2, for example, +-2 isotope errors are not counted as false. Beta version. (default: ‘0’) |
| FD:use_RNA_averagine | If set, RNA averagine model is used. |
| FD:preceding_MS1_count |
Specifies the number of preceding MS1 spectra for MS2 precursor determination. In TDP, the precursor peak of a MS2 spectrum may not belong to any deconvolved masses in the MS1 spectrum immediately preceding the MS2 spectrum. Increasing this parameter to N allows for the search for the deconvolved masses in the N preceding MS1 spectra from the MS2 spectrum, increasing the chance that its precursor is deconvolved. (default: ‘3’) (min: ‘1’) |
| FD:isolation_window |
Default isolation window with. If the input mzML file does not contain isolation window width information, this width will be used. (default: ‘5.0’) |
| FD:forced_MS_level |
If set to an integer N, MS level of all spectra will be set to N regardless of original MS level. Useful when deconvolving datasets containing only MS2 spectra. (default: ‘0’) (min: ‘0’) |
| FD:merging_method |
Method for spectra merging before deconvolution. 0: No merging 1: Average gaussian method to perform moving gaussian averaging of spectra per MS level . Effective to increase proteoform ID sensitivity (in particular for Q-TOF datasets). 2: Block method to perform merging of all spectra into a single one per MS level (e.g., for NativeMS datasets). (default: ‘0’) (min: ‘0’, max: ‘2’) |
Spectral deconvolution parameters: (with prefix SD: )
| parameter | Description |
|---|---|
| SD:tol |
Ppm tolerance for MS1, 2, … (e.g., -tol 10.0 5.0 to specify 10.0 and 5.0ppm for MS1 and MS2, respectively) (default: ‘[10.0 10.0]') |
| SD:min_mass |
Minimum mass (Da) (default: ‘50.0’) |
| SD:max_mass |
Maximum mass (Da) (default: ‘1.0e05’) |
| SD:min_charge |
Minimum charge state for MS1 spectra (can be negative for negative mode) (default: ‘1’) |
| SD:max_charge |
Maximum charge state for MS1 spectra (can be negative for negative mode) (default: ‘100’) |
| SD:precursor_charge |
Charge state of the target precursor. All precursor charge is fixed to this value. This parameter is useful for targeted studies where MS2 spectra are generated from a fixed precursor (e.g.,Native-MS). (default: ‘0’) (min: ‘0’) |
| SD:precursor_mz |
Target precursor m/z value. This option must be used with -target_precursor_charge option. Otherwise, it will be ignored. If -precursor_charge option is used but this option is not used, the precursor m/z value written in MS2 spectra will be used by default.(default: ‘0.0’) (min: ‘0.0’) |
| SD:min_cos |
Cosine similarity thresholds between avg. and observed isotope pattern for MS1, 2, … (e.g., -min_cos 0.3 0.6 to specify 0.3 and 0.6 for MS1 and MS2, respectively) (default: ‘[0.85 0.85]') |
| SD:min_snr |
Minimum charge SNR (the SNR of the isotope pattern of a specific charge) thresholds for MS1, 2, … (e.g., -min_snr 1.0 0.6 to specify 1.0 and 0.6 for MS1 and MS2, respectively) (default: ‘[1.0 1.0]') |
| SD:max_qvalue |
Qvalue thresholds for MS1, 2, … Effective only when FDR estimation is active. (e.g., -max_qvalue 0.1 0.2 to specify 0.1 and 0.2 for MS1 and MS2, respectively) (default: ‘[1.0 1.0]') |
Feature tracing parameters: (with prefix ft: )
| parameter | Description |
|---|---|
| ft:mass_error_ppm |
Feature tracing mass ppm tolerance. When negative, MS1 tolerance for mass deconvolution will be used (e.g., 16 ppm is used when -SD:tol 16). (default:'-1.0’) |
| ft:quant_method |
Method of quantification for mass traces. For LC data ‘area’ is recommended, ‘median’ for direct injection data. ‘max_height’ simply uses the most intense peak in the trace. (default: ‘area’) (valid: ‘area’, ‘median’, ‘max_height’) |
| ft:min_sample_rate |
Minimum fraction of scans along the feature trace that must contain a peak. To raise feature detection sensitivity, lower this value close to 0. (default: ‘0.1’) |
| ft:min_trace_length |
Minimum expected length of a mass trace (in seconds). Only for MS1 (or minim um MS level in the dataset) feature tracing. For MSn, all traces are kept regardless of this value. (default: ‘10.0’) |
| ft:max_trace_length |
Maximum expected length of a mass trace (in seconds). Set to a negative value to disable maximal length check during mass trace detection. (default:'-1.0’) |
| ft:min_cos |
Cosine similarity threshold between avg. and observed isotope pattern. When negative, MS1 cosine threshold for mass deconvolution will be used (default: ‘-1.0’) |
Isobaric quantification parameters: (with prefix iq: )
| parameter | Description |
|---|---|
| iq:type |
Isobaric Quantitation method used in the experiment. (default: ‘none’) (valid: ‘none’, ‘itraq4plex’, ‘itraq8plex’, ‘tmt10plex’, ‘tmt11plex’, ‘tmt16plex’, ‘tmt18plex’, ‘tmt6plex’) |
| iq:isotope_correction |
Enable isotope correction (highly recommended). Note that you need to provide a correct isotope correction matrix otherwise the tool will fail or produce invalid results. (default: ‘true’) (valid: ‘true’, ‘false’) |
| iq:reporter_mz_tol |
M/z tolerance in Th from the expected position of reporter ion m/zs. (default: ‘2.0e-03’) |
Tagger parameters: (with prefix tagger: )
| parameter | Description |
|---|---|
| tagger:max_tag_count |
Maximum number of the tags per length (lengths set by -min_length and -max_length options). The tags with different amino acid combinations are all treated separately. E.g., TII, TIL, TLI, TLL are distinct tags even though they have the same mass differences. but are counted as four different tags. (default: ‘0’) (min: ‘0’) |
| tagger:min_length |
Minimum length of a tag. Each mass gap contributes to a single length (even if a mass gap is represented by multiple amino acids). (default: ‘4’) (min:‘3’ max: ‘30’) |
| -tagger:max_length |
Maximum length of a tag. Each mass gap contributes to a single length (even if a mass gap is represented by multiple amino acids). (default: ‘10’) (min: ‘3’ max: ‘30’) |
| tagger:flanking_mass_tol |
Flanking mass tolerance in Da. (default: ‘200.0’) |
| tagger:max_iso_error_count |
Maximum isotope error count per tag. (default: ‘0’) (min: ‘0’ max: ‘2’) |
| tagger:min_matched_aa |
Minimum number of amino acids in matched proteins, covered by tags. (default: ‘5’) |
| tagger:fasta |
Target protein sequence database against which tags will be matched. |
| tagger:out |
Tagger output file. |
Running FLASHDeconv with GUI
GUI command is found under [OpenMS path]/bin directory. From the bin directory, type
./FLASHDeconvWizard
And this window pops up.

From the “LC-MS files” menu you can select (possibly multiple) mzML files to analyze. The selected files are analyzed with the same parameter set.
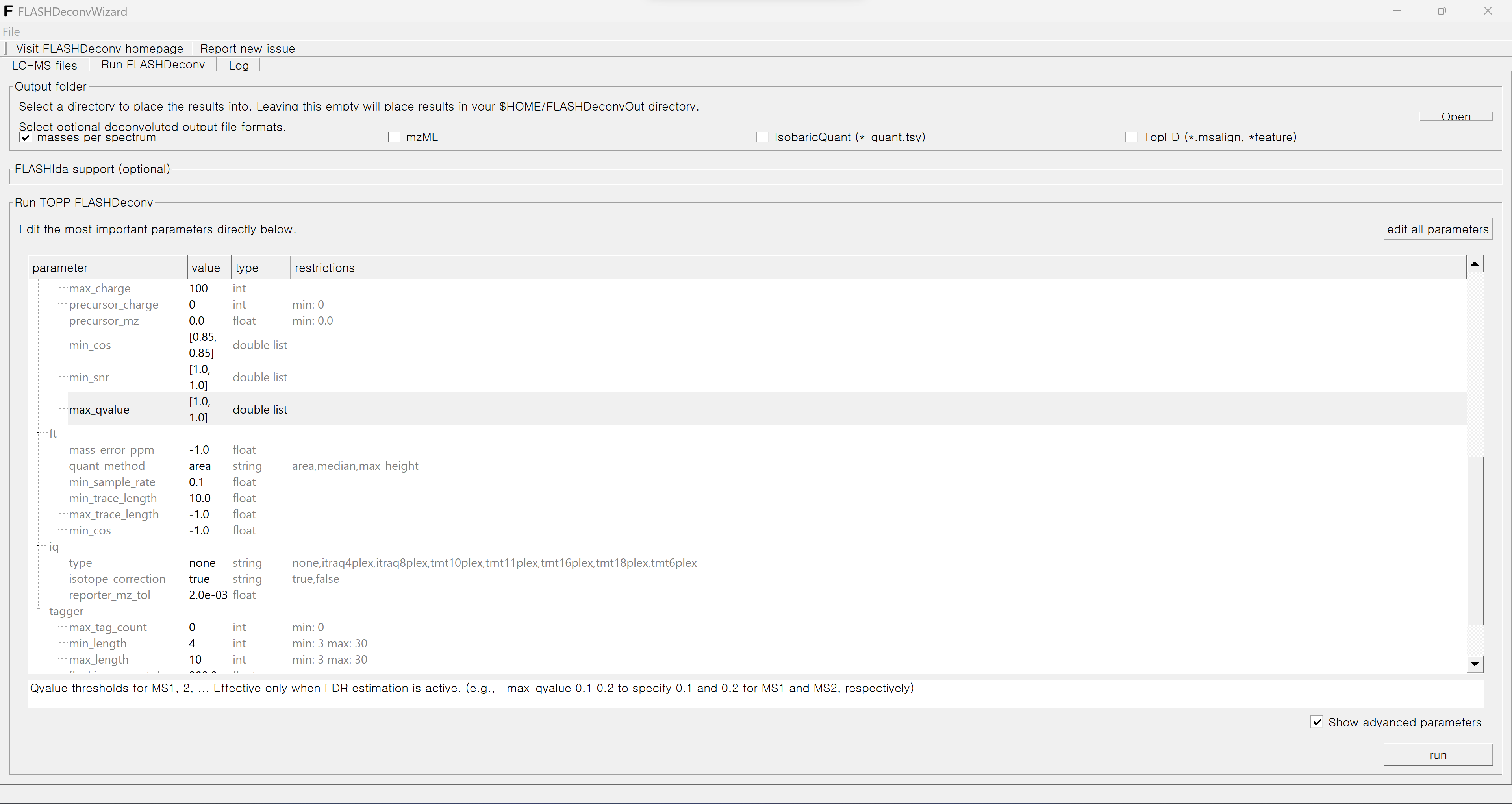
Then if you go to the “Run FLASHDeconv” menu, you can control all the parameters and output options.
You can see the progress in the log window.
The default output folder is [home directory]/FLASHDeconvOut folder. You may change this by using Browse button in the right side. Below we have four toggle output buttons.
If “masses per spectrum” is selected (selected by default), spectrum level deconvolution results (per MS level) are generated (in tsv format). In the command line, this is controlled with -out_spec option, and users must specify file name per MS level. But in GUI, simply activating this “masses per spectrum” button will set the output spectrum file name per MS level automatically.
If “mzML” is selected, the deconvolved spectra are generated in mzML format.
“Promex (.ms1ft)” triggers the Promex format output generation (only for MS1), and “TopFD (.msalign,*.feature)” triggers the TopFD format output generation (both msalign and feature formats). Again, these buttons override -out_promex, -out_topFD, and -out_topFD_feature in the command line and automatically set the output file names.
The box below the toggle buttons controls the parameters. In default it shows only basic parameters. If the “Show advanced parameters” toggle button is activated, the advanced parameters will appear.
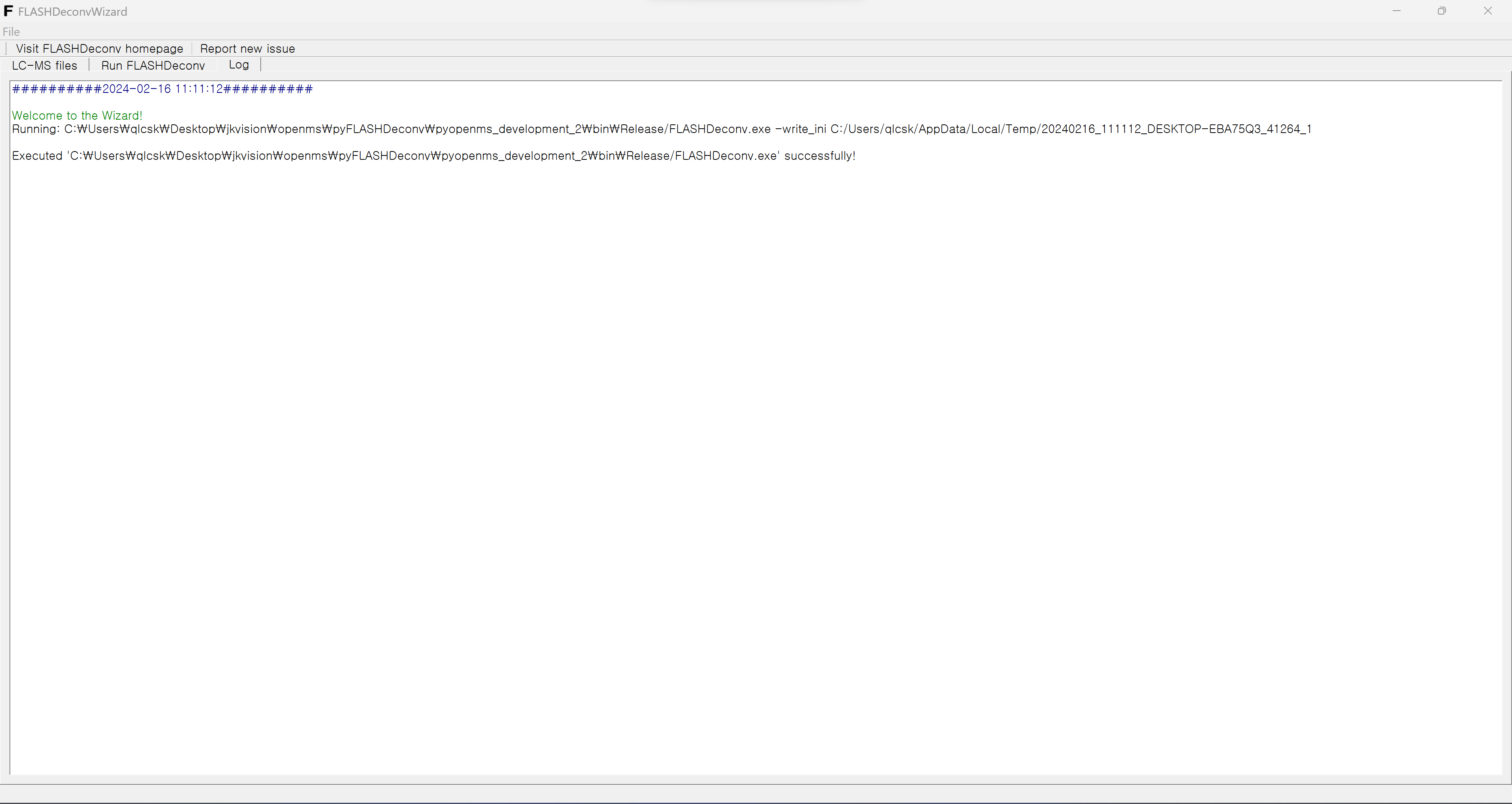
Lastly, the “Log” menu shows the log from FLASHDeconv. During or after FLASHDeconv run, one may check the log from FLASHDeconv from this menu. Here, also the command line commands corresponding to the current parameter selection by GUI also appear for reference and future use.
Running FLASHDeconv on command line
Runnable FLASHDeconv file can be found under [OpenMS path]/bin directory.
The mandatory options are -in and -out options. FLASHDeconv 2.0 only takes mzML file as its input. Basic parameters could be adjusted by the user according to instrumental setup. For input mzML file conversion from raw file, we recommend to use MSConvert with vendor provided peak picking methods.
For example if one wants to deconvolve /User/me/data/infile.mzml and get the result /User/me/out/outfilefeature.tsv,
one could run FLASHDeconv by typing as follows in the directory where FLASHDeconv is installed.
./FLASHDeconv -in /User/me/data/infile.mzml -out /User/me/out/outfilefeature.tsv
Output files
- Deconvolved feature file (*.tsv) specified by -out
- (optional) Deconvolved MSn spectra files (*.tsv) specified by -out_spec
- (optional) Deconvolved mzML spectra file (*.mzML) specified by -out_mzml
- (optional) Deconvolved MS1 in promex output format (*.ms1ft) specified by -out_promex
- (optional) Deconvolved MSn spectra files in topfd output format (*.msalign) specified by -out_topFD
- (optional) Deconvolved MSn feature files in topfd output format (*.feature) specified by -out_topFD_feature
Example datasets
Mass spectrometry datasets(*.raw and *.mzML) and corresponding results have been uploaded to MassIVE (https://massive.ucsd.edu) and are available under accession number MSV000084001.